Design concepts and requirements ¶
This document aims to describe the design requirements and principles of the dataframe interchange protcol, and the functionality it needs to support.
Conceptual model of a dataframe ¶
For a protocol to exchange dataframes between libraries, we need both a model of what we mean by “dataframe” conceptually for the purposes of the protocol, and a model of how the data is represented in memory:
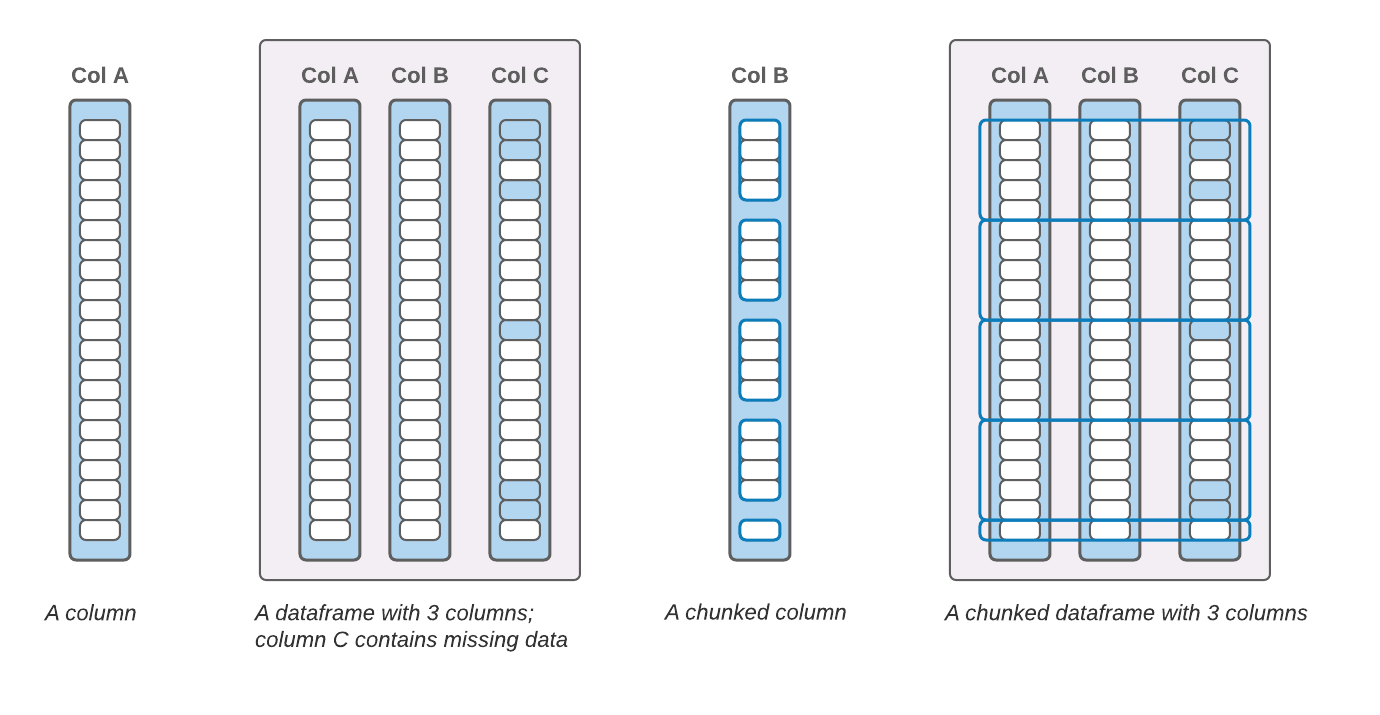
The smallest building blocks are 1-D arrays (or “buffers”), which are contiguous in memory and contain data with the same dtype. A column consists of one or more 1-D arrays (if, e.g., missing data is represented with a boolean mask, that’s a separate array). A dataframe contains one or more columns. A column or a dataframe can be “chunked”; a chunk is a subset of a column or dataframe that contains a set of (neighboring) rows.
Protocol design requirements ¶
-
Must be a standard Python-level API that is unambiguously specified, and not rely on implementation details of any particular dataframe library.
-
Must treat dataframes as an ordered collection of columns (which are conceptually 1-D arrays with a dtype and missing data support).
Note: this relates to the API for
__dataframe__, and does not imply that the underlying implementation must use columnar storage! -
Must allow the consumer to select a specific set of columns for conversion.
-
Must allow the consumer to access the following “metadata” of the dataframe: number of rows, number of columns, column names, column data types.
Note: this implies that a data type specification needs to be created. Note: column names are required; they must be strings and unique. If a dataframe doesn’t have them, dummy ones like
'0', '1', ...can be used. -
Must include device support.
-
Must avoid device transfers by default (e.g. copy data from GPU to CPU), and provide an explicit way to force such transfers (e.g. a
force=orcopy=keyword that the caller can set toTrue). -
Must be zero-copy wherever possible.
-
Must support missing values (
NA) for all supported dtypes. -
Must support string, categorical and datetime dtypes.
-
Must allow the consumer to inspect the representation for missing values that the producer uses for each column or data type. Sentinel values, bit masks, and boolean masks must be supported. Must also be able to define whether the semantic meaning of
NaNandNaTis “not-a-number/datetime” or “missing”.Rationale: this enables the consumer to control how conversion happens, for example if the producer uses
-128as a sentinel value in anint8column while the consumer uses a separate bit mask, that information allows the consumer to make this mapping. -
Must allow the producer to describe its memory layout in sufficient detail. In particular, for missing data and data types that may have multiple in-memory representations (e.g., categorical), those representations must all be describable in order to let the consumer map that to the representation it uses.
Rationale: prescribing a single in-memory representation in this protocol would lead to unnecessary copies being made if that represention isn’t the native one a library uses.
Note: the memory layout is columnar. Row-major dataframes can use this protocol, but not in a zero-copy fashion (see requirement 2 above).
-
Must support chunking, i.e. accessing the data in “batches” of rows. There must be metadata the consumer can access to learn in how many chunks the data is stored. The consumer may also convert the data in more chunks than it is stored in, i.e. it can ask the producer to slice its columns to shorter length. That request may not be such that it would force the producer to concatenate data that is already stored in separate chunks.
Rationale: support for chunking is more efficient for libraries that natively store chunks, and it is needed for dataframes that do not fit in memory (e.g. dataframes stored on disk or lazily evaluated).
-
May (desired, not required) support
__dlpack__as the array interchange protocol at the individual buffer level for dtypes that are supported by DLPack.Rationale: there is a connection between dataframe and array interchange protocols. If we treat a dataframe as a set of columns which each are a set of 1-D arrays (there may be more than one in the case of using masks for missing data, or in the future for nested dtypes), it may be expected that there is a connection to be made with the array data interchange method. The array interchange is based on DLPack; its major limitation from the point of view of dataframes is the lack of support of all required data types (string, categorical, datetime) and missing data.
We’ll also list some things that were discussed but are not requirements:
-
Object dtype does not need to be supported
-
Nested/structured dtypes within a single column does not need to be supported.
Rationale: not used a lot, additional design complexity not justified. May be added in the future (does have support in the Arrow C Data Interface). Also note that Arrow and NumPy structured dtypes have different memory layouts, e.g. a
(float, int)dtype would be stored as two separate child arrays in Arrow and as a singlef0, i0, f1, i1, ...interleaved array in NumPy. -
Extension dtypes, i.e. a way to extend the set of dtypes that is explicitly supported, are out of scope.
Rationale: complex to support, not used enough to justify that complexity.
-
Support for strided storage in buffers.
Rationale: this is supported by a subset of dataframes only, mainly those that use NumPy arrays. In many real-world use cases, strided arrays will force a copy at some point, so requiring contiguous memory layout (and hence an extra copy at the moment
__dataframe__is used) is considered a good trade-off for reduced implementation complexity. -
“virtual columns”, i.e. columns for which the data is not yet in memory because it uses lazy evaluation, are not supported other than through letting the producer materialize the data in memory when the consumer calls
__dataframe__.Rationale: the full dataframe API will support this use case by “programming to an interface”; this data interchange protocol is fundamentally built around describing data in memory .
To be decided ¶
Should there be a standard
from_dataframe
constructor function?
This
isn’t completely necessary, however it’s expected that a full dataframe API
standard will have such a function. The array API standard also has such a
function, namely
from_dlpack
. Adding at least a recommendation on syntax
for this function makes sense, e.g., simply
from_dataframe(df)
.
Discussion at
dataframe-api/issues/29
is relevant.
Frequently asked questions ¶
Can the Arrow C Data Interface be used for this? ¶
What we are aiming for is quite similar to the Arrow C Data Interface (see
the
rationale for the Arrow C Data Interface
),
except
__dataframe__
is a Python-level rather than C-level interface.
The data types format specification of that interface is something that could
be used unchanged.
The main limitation seems to be that Arrow does not have device support
–
@kkraus14
will bring this up on the Arrow dev mailing list. Another
identified issue is that the “deleter” on the Arrow C struct is present at the
column level, and there are use cases for having it at the buffer level
(mixed-device dataframes, more granular control over memory).
Note that categoricals are supported, Arrow uses the phrasing “dictionary-encoded types” for categorical. Also, what it calls “array” means “column” in the terminology of this document (and every Python dataframe library).
The Arrow C Data Interface says specifically it was inspired by
Python’s
buffer protocol
, which is also
a C-only and CPU-only interface. See
__array_interface__
below for a
Python-level equivalent of the buffer protocol.
Note that specifying the precise semantics for implementers (both producing and consuming libraries) will be important. The Arrow C Data interface relies on providing a deletion / finalization method similar to DLPack. The desired semantics here need to be ironed out. See Arrow docs on release callback semantics .
Is
__dataframe__
analogous to
__array__
or
__array_interface__
?
¶
Yes, it is fairly analogous to
__array_interface__
. There will be some
differences though, for example
__array_interface__
doesn’t know about
devices, and it’s a
dict
with a pointer to memory so there’s an assumption
that the data lives in CPU memory (which may not be true, e.g. in the case of
cuDF or Vaex).
It is
not
analogous to
__array__
, which is NumPy-specific.
__array__
is a
method attached to array/tensor-like objects, and calling it is requesting
the object it’s attached to to turn itself into a NumPy array. Hence, the
library that implements
__array__
must depend (optionally at least) on
NumPy, and call a NumPy
ndarray
constructor itself from within
__array__
.
What is wrong with
.to_numpy?
and
.to_arrow()
?
¶
Such methods ask the object it is attached to to turn itself into a NumPy or Arrow array. Which means each library must have at least an optional dependency on NumPy and on Arrow if it implements those methods. This leads to unnecessary coupling between libraries, and hence is a suboptimal choice - we’d like to avoid this if we can.
Instead, it should be dataframe consumers that rely on NumPy or Arrow, since
they are the ones that need such a particular format. So, it can call the
constructor it needs. For example,
x
=
np.asarray(df['colname'])
(where
df
supports
__dataframe__
).
A related question is: can
__array__
and/or
__arrow_array__
be used at the
column level? This is more reasonable, but probably does lead to more
complexity for very limited gains - for an issue with discussion on that, see
dataframe-api/issues/48
.
Does an interface describing memory work for virtual columns? ¶
Vaex is an example of a library that can have “virtual columns” (see
@maartenbreddels
comment here
).
If the protocol includes a description of data layout in memory, does that
work for such a virtual column?
Yes. Virtual columns need to be materialized in memory before they can be
turned into a column for a different type of dataframe - that will be true
for every discussed form of the protocol; whether there’s a
to_arrow()
or
something else does not matter. Vaex can choose
how
to materialize (e.g.,
to an Arrow array, a NumPy array, or a raw memory buffer) - as long as the
returned description of memory layout is valid, all those options can later
be turned into the desired column format without a data copy, so the
implementation choice here really doesn’t matter much.
Note: the above statement on materialization assumes that there are many
forms a virtual column can be implemented, and that those are all
custom/different and that at this point it makes little sense to standardize
that. For example, one could do this with a simple string DSL (
'col_C
=
col_A
+
col_B'
, with a fancier C++-style lazy evaluation, with a
computational graph approach like Dask uses, etc.).
Possible direction for implementation ¶
Rough initial prototypes (historical) ¶
The
cuDFDataFrame
,
cuDFColumn
and
cuDFBuffer
sketched out by @kkraus14
here
looked like it was in the right direction.
This prototype by Wes McKinney was the first attempt, and has some useful features.
Relevant existing protocols ¶
Here are the four most relevant existing protocols, and what requirements they support:
| supports | buffer protocol |
__array_interface__
|
DLPack | Arrow C Data Interface |
|---|---|---|---|---|
| Python API | Y | (1) | ||
| C API | Y | Y | Y | Y |
| arrays | Y | Y | Y | Y |
| dataframes | ||||
| chunking | ||||
| devices | Y | |||
| bool/int/uint/float | Y | Y | Y | Y |
| missing data | (2) | (3) | (4) | Y |
| string dtype | (4) | (4) | Y | |
| datetime dtypes | (5) | Y | ||
| categoricals | (6) | (6) | (7) | Y |
-
The Python API is only an interface to call the C API under the hood, it doesn’t contain a description of how the data is laid out in memory.
-
Can be done only via separate masks of boolean arrays.
-
__array_interface__has amaskattribute, which is a separate boolean array also implementing the__array_interface__protocol. -
Only fixed-length strings as sequence of char or unicode.
-
Only NumPy datetime and timedelta, not timezones. For the purpose of data interchange, timezones could be represented separately in metadata if desired.
-
No explicit support, however categoricals can be mapped to either integers or strings. Unclear how to communicate that information from producer to consumer.
-
No explicit support, categoricals can only be mapped to integers.
| implementation | buffer protocol |
__array_interface__
|
DLPack | Arrow C Data Interface |
|---|---|---|---|---|
| refcounting | Y | Y | ||
| call deleter | Y | Y | ||
| supporting C code | in CPython | in NumPy | spec-only | spec-only |
It is worth noting that for all four protocols, both dataframes and chunking can be easily layered on top. However only arrays, which are the only parts that have a specific memory layout, are explicitly specified in all those protocols. For Arrow this would be a little easier than for other protocols given the inclusion of “children” and “dictionary-encoded types”, and indeed PyArrow already does provide such functionality.
Data type descriptions ¶
There are multiple options for how to specify the dtype. The buffer protocol, NumPy and Arrow use format strings, DLPack uses enums. Furthermore dtype literals can be used for a Python API; this is what the array API standard does. Here are some examples:
| dtype | buffer protocol |
__array_interface__
|
DLPack | Arrow C Data Interface |
|---|---|---|---|---|
| int8 |
'=b'
|
'i1'
|
(0, 8)
|
'c'
|
| int32 |
=i'
|
'i4'
|
(0, 32)
|
'i'
|
| float64 |
'=d'
|
'<f8'
|
(2, 64)
|
'g'
|
| utf-8 string |
'u'
or
'w'
|
'U'
|
n.a. |
'u'
|
The
=
,
<
,
>
are denoting endianness; Arrow only supports native endianness.